New tool explores biological differences and new targeted therapies in childhood acute lymphoblastic leukemia
Researchers at the Department of Oncology-Pathology have published a paper in Nature Communications where they created a cell line resource dataset and analysis tool that could be used to identify the traits that make specific leukemias sensitive to new types of targeted therapies.
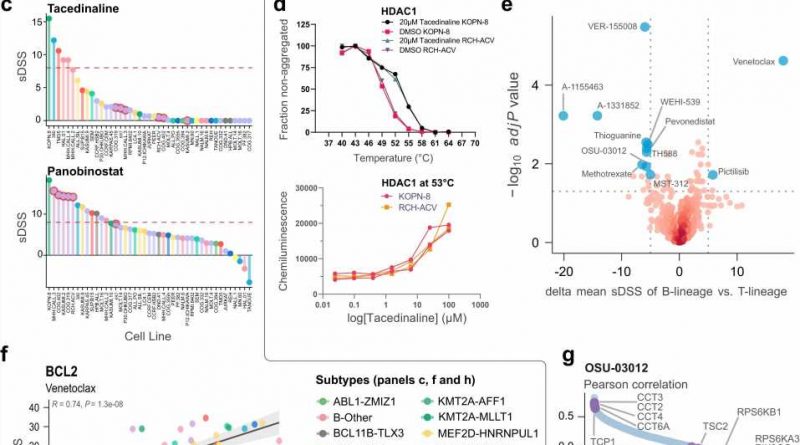
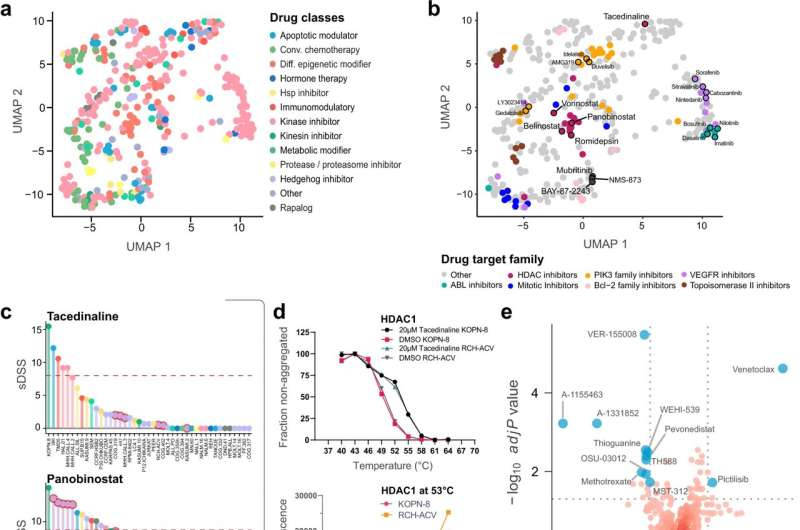
The researchers used cell lines from childhood acute lymphoblastic leukemia (ALL) patients with different forms of the disease and analyzed proteins, RNAs, and sensitivity to 528 drugs clinically approved or under investigation. Comparing cell lines that responded to drugs in different ways could identify drug mechanisms and the traits that make leukemias sensitive or resistant to those therapies. For some drugs, the researchers identified that they might work through secondary off-target mechanisms. In a higher-risk ALL subtype, which responds poorly to standard chemotherapy, they identified unexpected sensitivity to a type of immunotherapy drug, which increases the strength of the molecular signals that activate immune cells.
ALL is the most common childhood cancer. Most patients can be treated with chemotherapy and stem cell transplantation, but 15–20% will respond poorly, and this patient group lacks treatment options and has high mortality risk. Additionally, current standard treatments can have a long-term health burden for children who survive their cancer. There is a need to identify therapeutics which are less severe than standard treatments and which could help patients that don’t respond to current options. The researchers’ results provide a foundation for exploring biological differences and new targeted therapies in childhood ALL, and serve as a broad public resource that will support other research projects.
The researchers used a panel of 49 childhood ALL cell lines and acquired mass-spectrometry based in-depth proteome measurements, RNA sequencing, high-throughput drug screening, and fusion gene analysis. They created a web-based platform which allows the public to explore and analyze the dataset. They performed analyses that integrated these data types and improved understanding of biological differences. They performed analyses to profile the mechanisms of effective drugs in the screen. They performed validation experiments to understand drug mechanisms in depth.
Source: Read Full Article