Is your body out of sync? Study finds organs age at varying rates
In a recent study published in the journal Nature, researchers used cutting-edge blood plasma proteomics to investigate if human organs age at different rates. They analyzed 11 organs in almost 5,700 adults of different ages and found that nearly 20% of study participants experienced accelerated organ aging in at least one organ. Alarmingly, 1.7% of participants depicted accelerated aging in multiple organs. They followed these findings with an estimation of the potential increases in risk of age-related diseases. Their results present that accelerated organ aging is associated with a 250% increased risk of cardiac failure and a heightened risk of Alzheimer's disease.
 Study: Organ aging signatures in the plasma proteome track health and disease. Image Credit: Icruci / Shutterstock
Study: Organ aging signatures in the plasma proteome track health and disease. Image Credit: Icruci / Shutterstock
Is age really just a number?
Aging is a universally detrimental process resulting in the deterioration of the structure and function of somatic tissues. Since natural selection is blind to all non-reproductive-success-related diseases, aging beyond reproductive age is associated with a drastic increase in non-communicable conditions, including cardiovascular diseases (CVDs), cognitive impairment (such as Alzheimer's disease), and cancers.
Extensive studies on animal systems, especially murine models, have revealed molecular changes across multiple mouse organs, which, in turn, have been found to result in brain, heart, and kidney diseases. Unexpectedly, these studies revealed discordance between animal (mouse) age and organ age, with the same mouse presenting differences in aging-associated biomarkers across its organs.
Studies on human aging, while available, are scarce and share a common demerit of using magnetic resonance imaging (MRI)-based analyses. Unfortunately, MRI systems are restricted to being capable of measuring only brain volume and functional connectivity; they fail to establish the molecular underpinnings of observed results. Clinical chemistry approaches have attempted to bridge the MRI-associated knowledge gap, but the biomarkers used herein depict low organ specificity and hard susceptibility to bias and error.
In recent years, blood plasma biomarkers have increasingly provided an ideal means of accurate molecular aging of mice models, but this approach has yet to be applied to human subjects.
"A molecular understanding of human organ aging is of critical importance to address the massive global disease burden of aging and could revolutionize patient care, preventative medicine and drug development."
About the study
In the present study, researchers used blood plasma from 5,676 participants across five distinct study cohorts to discover and map a human organ-specific plasma proteome. They identified and measured 4,979 proteins, which were then used to develop and train models of organ aging. Organ-enriched proteins are characterized by having four times or greater protein abundance when compared to other organs. Of the 4.969 proteins analyzed, 19% (893) proteins were found to be enriched and were used for modeling and analyses.
A bagged ensemble of least absolute shrinkage and selection operator (LASSO) machine learning (ML) model was trained to identify organ-specific aging. The model was optimized to evaluate the age of 11 major organ types: adipose tissue, brain, artery, heart, immune tissue, kidney, intestine, lung, liver, pancreas, and muscles. These organs were chosen due to previous research which has associated these systems with age-related mortality and morbidity. Additionally, 3,907 non-enriched proteins were used to train an 'organismal' ML model, and all 4,979 proteins were used to elucidate the global effects of organ aging.
Data from two of the five cohorts was used to investigate the association between organ age and disease risk. Hazard ratios (HRs) for mortality and morbidity were computed. Finally, a separate 'second-generation brain aging model' termed CognitionBrain was developed using only the brain-associated enriched proteins to elucidate the impacts of brain aging on future cognitive performance.
Study findings
This study presents the first investigation to determine human organ-specific aging using molecular rather than conventional MRI approaches. Proteomic analyses using next-generation sequencing revealed more than 4700 proteins associated with organ-specific aging, 18% of which were enriched only in a single organ, thereby highlighting their potential as future organ-age biomarkers.
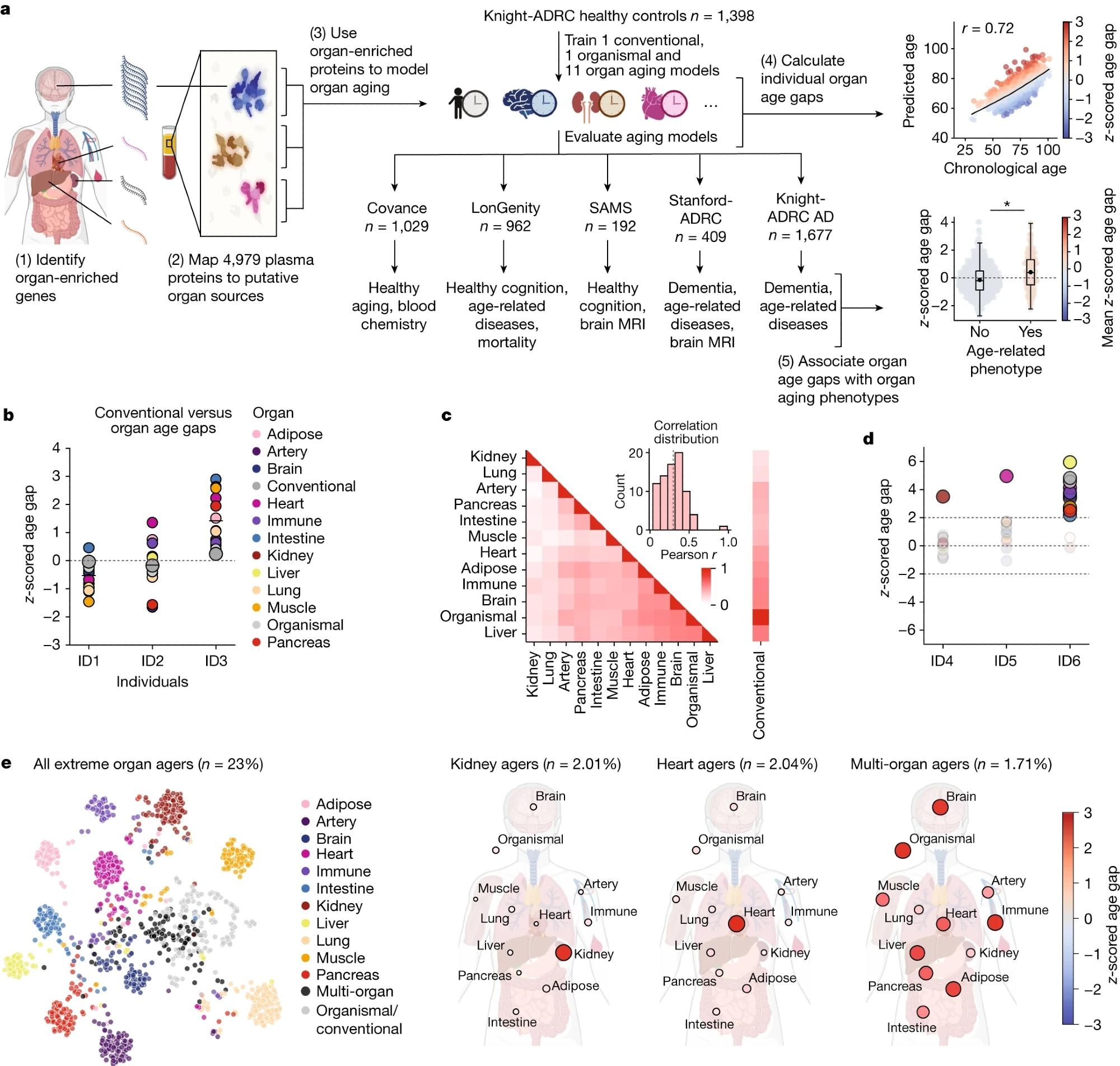 a, Study design to estimate organ-specific biological age. A gene was called organ-specific if its expression was four-fold higher in one organ compared to any other organ in GTEX bulk organ RNA-seq. This annotation was then mapped to the plasma proteome. Mutually exclusive organ-specific protein sets were used to train bagged LASSO chronological age predictors with data from 1,398 healthy individuals in the Knight-ADRC cohort. An ‘organismal’ model, which used the nonorgan-specific (organ shared) proteins, and a ‘conventional’ model, which used all proteins regardless of specificity, were also trained. Models were tested in four independent cohorts: Covance (n = 1,029), LonGenity (n = 962), SAMS (n = 192) and Stanford-ADRC (n = 420); models were also tested in the AD patients in the Knight-ADRC cohort (n = 1,677). To test the validity of organ aging models, the age gap was associated with multiple measures of health and disease. An example age prediction (predicted versus chronological age) and an example age gap versus phenotype association (age gap versus phenotype, standard boxplot) are shown. b, Individuals (ID) with the same conventional age gap can have different organ age gap profiles. Three example participants are shown. Bar represents mean age gap across n = 13 age gaps. c, Pairwise correlation of organ age gaps from n = 3,774 healthy participants across all cohorts. Distribution of all pairwise correlations is shown in inset histogram, with dotted line median correlation. The control age gap was highly correlated with the organismal age gap (r = 0.98), the sole outlier in the inset distribution plot. d, Identification of extreme agers, defined by a two standard deviation increase or decrease in at least one age gap. A representative kidney ager, heart ager and multi-organ ager are shown. e, All extreme agers were identified (23% of all n = 5,676 individuals) and clustered after setting age gaps below an absolute z-score of 2 to 0. The mean age gaps for all organs in the kidney agers, heart agers and multi-organ agers clusters are shown.
a, Study design to estimate organ-specific biological age. A gene was called organ-specific if its expression was four-fold higher in one organ compared to any other organ in GTEX bulk organ RNA-seq. This annotation was then mapped to the plasma proteome. Mutually exclusive organ-specific protein sets were used to train bagged LASSO chronological age predictors with data from 1,398 healthy individuals in the Knight-ADRC cohort. An ‘organismal’ model, which used the nonorgan-specific (organ shared) proteins, and a ‘conventional’ model, which used all proteins regardless of specificity, were also trained. Models were tested in four independent cohorts: Covance (n = 1,029), LonGenity (n = 962), SAMS (n = 192) and Stanford-ADRC (n = 420); models were also tested in the AD patients in the Knight-ADRC cohort (n = 1,677). To test the validity of organ aging models, the age gap was associated with multiple measures of health and disease. An example age prediction (predicted versus chronological age) and an example age gap versus phenotype association (age gap versus phenotype, standard boxplot) are shown. b, Individuals (ID) with the same conventional age gap can have different organ age gap profiles. Three example participants are shown. Bar represents mean age gap across n = 13 age gaps. c, Pairwise correlation of organ age gaps from n = 3,774 healthy participants across all cohorts. Distribution of all pairwise correlations is shown in inset histogram, with dotted line median correlation. The control age gap was highly correlated with the organismal age gap (r = 0.98), the sole outlier in the inset distribution plot. d, Identification of extreme agers, defined by a two standard deviation increase or decrease in at least one age gap. A representative kidney ager, heart ager and multi-organ ager are shown. e, All extreme agers were identified (23% of all n = 5,676 individuals) and clustered after setting age gaps below an absolute z-score of 2 to 0. The mean age gaps for all organs in the kidney agers, heart agers and multi-organ agers clusters are shown.
Results of three separate ML algorithms reveal that organ-specific aging was prevalent in 20% of the nearly 6,000 individuals sampled. The aging of specific organs, most notably the kidneys and heart, was associated with a significantly (~250%) increased risk of future comorbidities. Analyses of brain aging revealed substantial cognitive reduction (immediate) along with a significantly higher risk of developing memory and mental disorders, including Alzheimer's disease, in participants with accelerated brain growth.
"There are many future directions for this work. While we have shown that plasma proteomic organ aging models are distinct from previous proteomics models, clinical chemistry-based models and imaging-based models, future studies should assess how proteomic organ aging relates to other molecular measures of aging and disease such as methylation aging clocks and disease-specific prediction models."
Conclusions
In the first study of its kind conducted on humans, researchers used blood plasma proteomics to elucidate the protein biomarkers and molecular basis of organ-specific aging. Their analyses of almost 6,000 participants across five distinct study cohorts revealed over 4,700 proteins associated with early organ aging, of which 18% were organ-specific and could be used both for ML model training and as future diagnostic biomarkers.
Their findings revealed that a staggering 20% of participants experienced early aging in at least one organ, with almost 2% presenting multiple organ age acceleration. Organ aging was found to significantly increase mortality and cognitive risk, with the kidneys, CVD, and brain displaying the most detrimental effects.
"…we show that large-scale plasma proteomics and machine learning can be leveraged to noninvasively measure organ health and aging in living people. We show that biologically motivated modelling, in which we use sets of organ-specific proteins and the FIBA algorithm to further subset to physiological age-related proteins, enables deconvolution of the different rates of aging within an individual and measurement of aging at organ-level resolution."
- Oh, Hamilton S., et al. "Organ Aging Signatures in the Plasma Proteome Track Health and Disease." Nature, vol. 624, no. 7990, 2023, pp. 164-172, https://doi.org/10.1038/s41586-023-06802-1, https://www.nature.com/articles/s41586-023-06802-1
Posted in: Men's Health News | Medical Science News | Medical Research News | Women's Health News
Tags: Adipose, Aging, Alzheimer's Disease, Blood, Brain, Diagnostic, Gene, Heart, Imaging, Kidney, Liver, Machine Learning, Magnetic Resonance Imaging, Medicine, Mortality, Pancreas, Phenotype, Protein, Proteome, Proteomics, Research, RNA

Written by
Hugo Francisco de Souza
Hugo Francisco de Souza is a scientific writer based in Bangalore, Karnataka, India. His academic passions lie in biogeography, evolutionary biology, and herpetology. He is currently pursuing his Ph.D. from the Centre for Ecological Sciences, Indian Institute of Science, where he studies the origins, dispersal, and speciation of wetland-associated snakes. Hugo has received, amongst others, the DST-INSPIRE fellowship for his doctoral research and the Gold Medal from Pondicherry University for academic excellence during his Masters. His research has been published in high-impact peer-reviewed journals, including PLOS Neglected Tropical Diseases and Systematic Biology. When not working or writing, Hugo can be found consuming copious amounts of anime and manga, composing and making music with his bass guitar, shredding trails on his MTB, playing video games (he prefers the term ‘gaming’), or tinkering with all things tech.



