Comprehensive blood atlas of COVID-19 identifies hallmarks of disease severity
Researchers in the UK have presented a multi-omics integrated blood atlas that delineates the host immune response in patients with coronavirus disease 2019 (COVID-19) of varying severity, providing a unique reference resource for the interpretation of datasets generated from interventional trials.
Through the COvid-19 Multi-omics Blood ATlas (COMBAT) consortium, David Ahern from the University of Oxford and colleagues characterized COVID-19 of varying severity and identified immune signatures and correlates of host response.
The team says that integrative approaches such as those applied here are essential to better differentiating COVID-19 patients according to disease severity, underlying pathophysiology and infectious etiology.
“Our systems-based integrative approach and blood atlas will inform future drug development, clinical trial design and personalized medicine approaches for COVID-19,” write the researchers.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes peer review.
.jpg)
COVID-19 pathophysiology is highly heterogeneous between patients
The pathophysiology induced by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes COVID-19 – involves a complex interplay between lung pathology and maladaptive immune system responses.
Severe COVID-19 is characterized by hypoxia and can progress to acute respiratory distress syndrome, multiple organ failure and death.
Predisposing factors for severe disease include age, gender, ethnicity, obesity and comorbidities.
Currently, targeted precision treatment approaches are limited by an incomplete understanding of the clinical heterogeneity among patients and of the potentially druggable immune mediators of disease.
What did the researchers do?
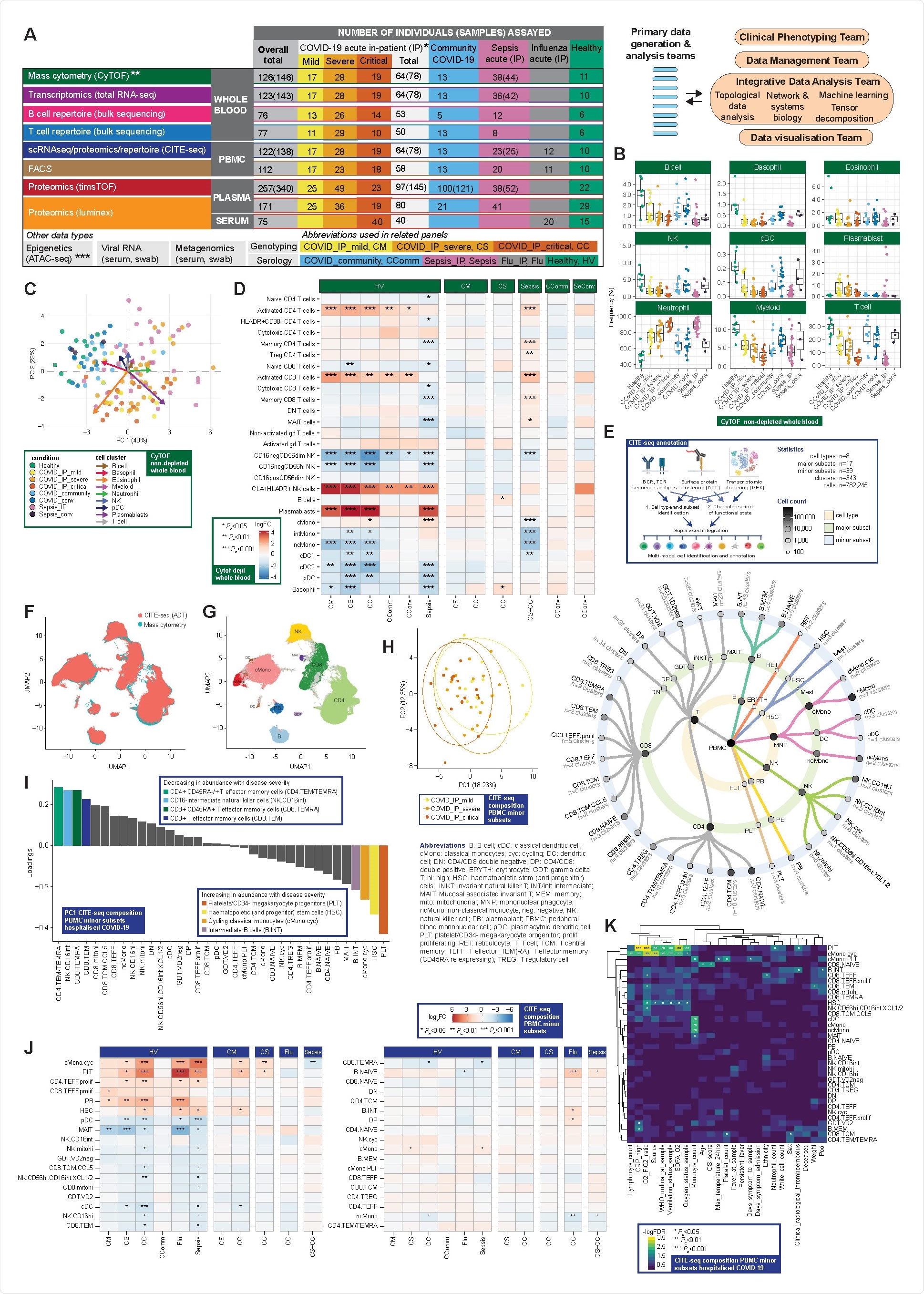
Now, through the (COMBAT) consortium, Ahern and colleagues have used a comprehensive multi-omics blood atlas in patients with varying COVID-19 severity to identify cells, mediators and pathways that are hallmarks of increasing disease severity.
To characterize the peripheral blood response to COVID-19, the researchers analyzed a prospective cohort of adult patients with confirmed SARS-CoV-2 infection who presented to clinical services during February and March 2020 – at the start of the UK pandemic and prior to any implementation of approved treatments or vaccination.
The team recruited a prospective cohort of 116 patients hospitalized with COVID-19 at Oxford University Hospitals. Samples were collected during acute admission and from 28 days following hospital discharge of convalescent individuals.
Results from these samples were compared with those from recovering COVID-19 cases in the community who had not required hospitalization, age-matched healthy volunteers, influenza cases requiring intensive care, and all-cause sepsis patients.
What did they find?
The team’s comprehensive, multi-modal analysis of multiple well-defined patient cohorts and healthy volunteers identified blood hallmarks of COVID-19 severity and specificity involving particular immune cell populations, components of innate and adaptive immunity, and connectivity with the inflammatory response.
The team found evidence for increased levels of activated CD8+ T cells and natural killer cell populations in cases of COVID-19 and failed clonal expansion in CD8+ T effector and central memory cells as disease severity increased.
Proteomic analysis identified specific plasma cytokine and chemokine levels as biomarkers of severe disease, with evidence for the following as hallmarks: acute phase inflammation, complement activation, fibrin clots, proteases, serum amyloid, tissue necrosis, receptor-mediated endocytosis and cholesterol transport.
Plasma protein signatures could be used to stratify patients
The study revealed plasma protein signatures that can be used to stratify hospitalized patients with acute COVID-19 into disease sub-phenotypes, with cluster membership associated with differential 28-day mortality.
“Patient stratification is important given the observed clinical heterogeneity within severe COVID-19. Such variability has historically been a major confounder of clinical trials for targeted immune therapy in other severe infections,” writes the team.
The researchers also found that the ratio of integrin alpha 4 (CD49d) to leukosialin (CD43) is specifically elevated in COVID-19 patients, including into recovery and during convalescence.
Reduced CD43 expression has been shown to lead to neutrophil retention in the bloodstream and increased adherence to vessel walls, which may be linked to the enhanced thrombosis observed in COVID-19 patients.
The CD49d:CD43 ratio may be an informative index score for neutrophil activities specific to COVID-19, suggests Ahern and colleagues.
AP-1 p38 MAPK pathway upregulation was a specific feature of COVID-19
The analysis also identified upregulation of the AP-1 p38 MAPK pathway as a specific feature of COVID-19 disease across different immune subsets.
“Combined with evidence of proliferation and cytokine response in these populations, this supports systemic immune activation and proliferation as a hallmark specific to COVID-19,” write the researchers.
The team says this finding is in keeping with recent evidence that baricitinib improves the recovery of hospitalized patients. This drug has been shown to act upstream of AP-1 and to control macrophage inflammation and neutrophil recruitment in COVID-19.
What are the implications of the findings?
The researchers say the multi-omics integrated blood atlas presented here provides a unique reference resource for replication and meta-analysis for interpreting datasets generated from interventional trials and includes tools for direct visualization (https://mlv.combat.ox.ac.uk/).
“Integrative approaches such as we have applied here are essential to better differentiate COVID-19 patients according to disease severity, underlying pathophysiology and infectious etiology,” writes Ahern and colleagues.
“This will be important as we seek novel therapeutic targets and the opportunity for a precision medicine approach to treatment that is appropriately timed and targeted to those patients most likely to benefit from a particular intervention,” concludes the team.
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Knight JC, et al. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. medRxiv, 2021. doi: https://doi.org/10.1101/2021.05.11.21256877, https://www.medrxiv.org/content/10.1101/2021.05.11.21256877v1
Posted in: Child Health News | Men's Health News | Medical Research News | Medical Condition News | Women's Health News | Disease/Infection News
Tags: Acute Respiratory Distress Syndrome, Assay, Blood, Cell, Chemokine, Cholesterol, Clinical Trial, Coronavirus, Coronavirus Disease COVID-19, Cytokine, Cytometry, Hospital, Hypoxia, Immune Response, Immune System, Inflammation, Influenza, Intensive Care, Macrophage, Medicine, Mortality, Necrosis, Obesity, Pandemic, Pathology, Pathophysiology, Personalized Medicine, Proliferation, Protein, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Sepsis, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Thrombosis

Written by
Sally Robertson
Sally first developed an interest in medical communications when she took on the role of Journal Development Editor for BioMed Central (BMC), after having graduated with a degree in biomedical science from Greenwich University.
Source: Read Full Article